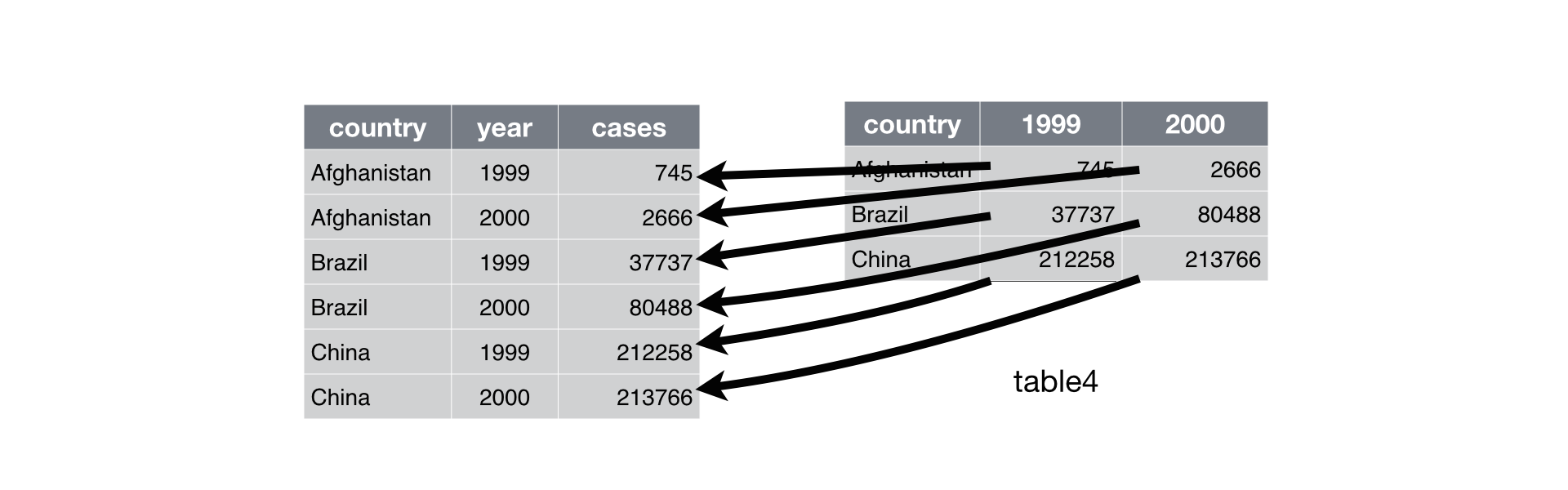
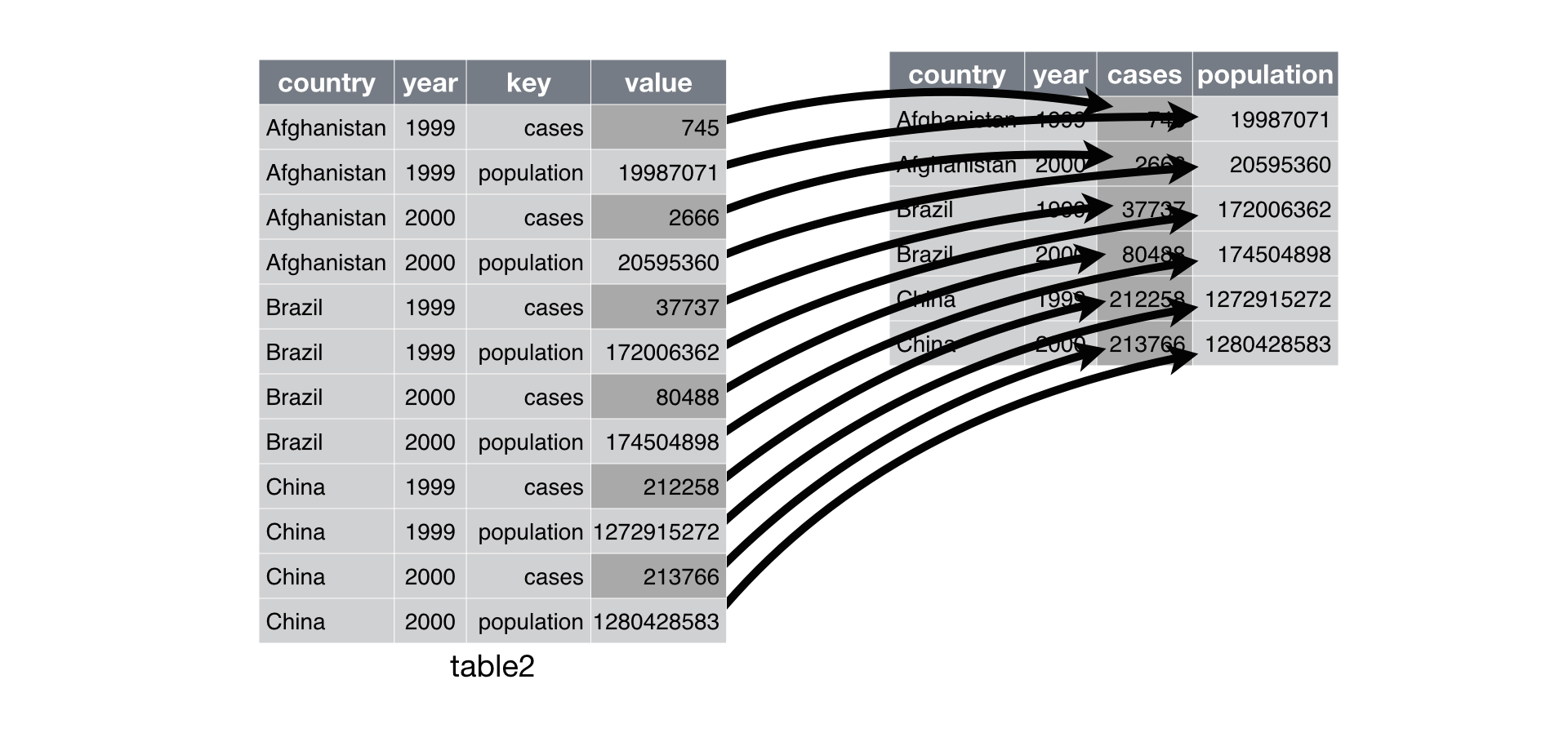
sessionInfo()R version 4.2.2 (2022-10-31)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur ... 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] digest_0.6.30 lifecycle_1.0.3 jsonlite_1.8.4 magrittr_2.0.3
[5] evaluate_0.18 rlang_1.0.6 stringi_1.7.8 cli_3.4.1
[9] rstudioapi_0.14 vctrs_0.5.1 rmarkdown_2.18 tools_4.2.2
[13] stringr_1.5.0 glue_1.6.2 htmlwidgets_1.6.0 xfun_0.35
[17] yaml_2.3.6 fastmap_1.1.0 compiler_4.2.2 htmltools_0.5.4
[21] knitr_1.41